Bridging the Gap
Comparing eDNA Metabarcoding with Traditional Survey Methods for Mammal Biodiversity Assessment
Mammals, a vital component of terrestrial ecosystems, are facing drastic declines due to anthropogenic changes in landscapes. With many species secluded in protected areas and facing extreme conservation status, there is a growing need for rapid, cost-effective, and noninvasive methods to monitor their populations. Within BCOMING, it is important that we use the most accurate techniques to establish biodiversity assessment as it is a core component in our research on pathogen circulation. Our team at Université de Liège in Belgium, consisting of Dr Pauline van Leeuwen and Prof. Johan Michaux, delves into the scientific literature for non-invasive methods to detect mammalian species in the field. They published the first scientific article supported by the BCOMING consortium. This study compares conventional survey methods and the emerging environmental DNA (eDNA) metabarcoding technique for mammal biodiversity assessment.
The Rise of eDNA Metabarcoding: Conventional survey methods, including trapping and field surveys, have been the cornerstone of mammal biodiversity assessment. However, the study highlights the emergence of eDNA metabarcoding as a high-throughput, multi-species detection tool based on PCR and Next-Generation Sequencing (NGS) technologies. This technique utilizes DNA extracted from environmental samples, providing a noninvasive means to monitor biodiversity. eDNA samples can be categorized into three groups based on substrates: eDNA from the environment (water, soil, sediments, and air filters), DNA traps (feces, hair, saliva, vegetation, even spider webs!), and invertebrate-derived DNA (leeches, flies, mosquitoes, dung beetles). Each substrate offers unique advantages for capturing genetic material, showcasing the versatility of eDNA metabarcoding.
Advantages and Challenges of eDNA Metabarcoding: Compared to traditional methods, eDNA metabarcoding presents several advantages, including reduced manpower, standardized collection across habitats, and noninvasiveness. However, challenges such as DNA degradation, PCR errors, and the need for extensive reference databases need to be addressed. The study emphasizes the importance of a nuanced approach and cross-validation with traditional methods.
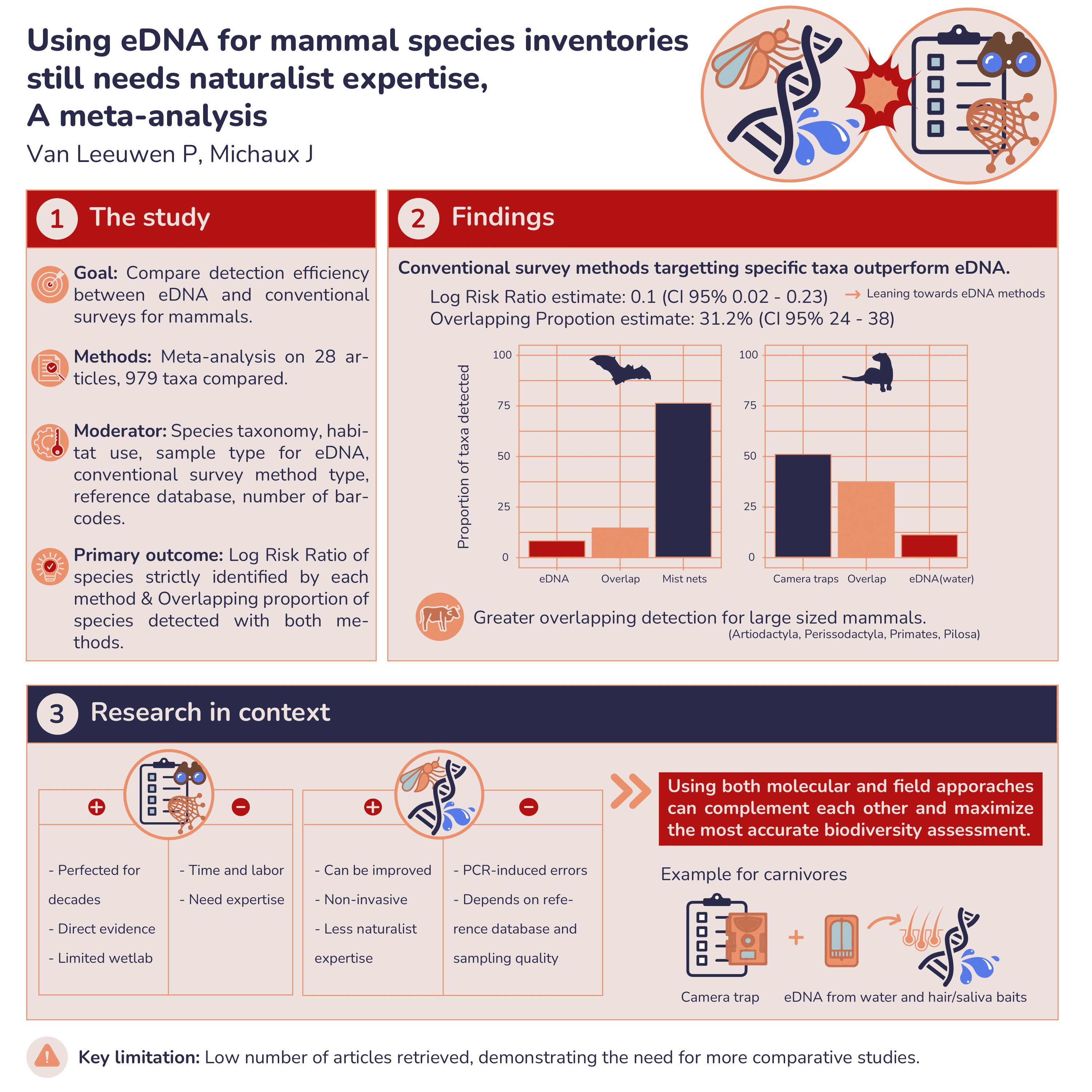
Taxonomy-Dependent Detection Success: A comprehensive meta-analysis, spanning from 2005 to 2023, reveals insights into the detection overlap between eDNA metabarcoding and traditional methods. Taxonomy, habitat use, sample type, traditional survey methods, reference databases, and the number of barcodes used are identified as moderators influencing detection success. Meta-regression further refines the analysis, providing a nuanced understanding of the factors affecting detection ratios.
The research explores the taxonomy-dependent success of eDNA metabarcoding, indicating variations in detection efficiency across mammalian orders. Carnivores exhibit higher detection success with traditional methods, especially camera traps, while eDNA performs well for ungulates and primates. The study highlights the need for tailored approaches based on the physiological and ecological characteristics of target species.
Overall, we underscore the promising potential of eDNA metabarcoding for mammal biodiversity assessment. However, we also emphasize the need for a hybrid approach, combining the strengths of both eDNA and traditional survey methods. Acknowledging the taxonomy-dependent nature of detection success, we call for a thoughtful integration of molecular tools and field expertise to maximize accuracy in biodiversity assessments. As eDNA continues to evolve, collaborative efforts between molecular biologists, taxonomists, and field ecologists are crucial for refining methodologies and expanding our understanding of mammalian populations.
For more information, here is the open access link to the publication: https://doi.org/10.1002/ece3.10788