Les interfaces zoonotiques sous la Loupe
Salmonella spp. chez les Chauves-Souris, les Rongeurs et les Chiens dans la région de Nan en Thaïlande.
Par Dr. Alix NICOLAS, Vétérinaire, dans le cadre de son stage de fin d’étude à l’Université de Liège, Belgique, et en partenariat avec Kasetsart University, Thaïlande pour le projet Spillover Interface
Durant mon stage de dernière année, j’ai eu l’opportunité de participer au projet BCOMING, en conduisant une recherche sur les interfaces zoonotiques sous la direction de mon promoteur de TFE (Travail de Fin d’Etude), le docteur Mutien Marie Garigliany du départements de pathologie de la facultée vétérinaire de Liège, de mon maître de stage le docteur Johan Michaux et avec la supervision du docteur Pauline Van Leeuwen du laboratoire de génétique de la conservation de la faculté de biologie de Liège. L'objectif principal de cette étude était de déterminer la prévalence de Salmonella spp. chez les chauves-souris, les rongeurs et les chiens dans la province de Nan en Thaïlande. Avec la récente pandémie de Covid-19, il est devenu évident que l'émergence de nouvelles maladies s'accélère. Les précédentes épidémies, telles que la grippe espagnole en 1918, la grippe aviaire en 2009-2010, ainsi que les différentes épidémies de coronavirus (SARS-CoV-1 en 2002-2003, MERS en 2012 et Covid-19 Jones et al., 2008; Smith et al., 2009), ont toutes pour point commun des sources animales.
Le projet BCOMING, financé par l'UE et soutenu par des collaborations internationales, vise à comprendre et prévenir l’émergence de nouvelles maladies infectieuses. En parallèle, le projet Spillover Interface a des objectifs similaires. Ils se concentrent sur le lien entre la perte de biodiversité et l'émergence de maladies zoonotiques, tout en cherchant à limiter leur transmission par des stratégies de conservation et des réseaux de surveillance efficaces. Ces projets explorent un gradient d’urbanisation, allant des écosystèmes les moins anthropisés aux plus urbains, ainsi qu’un gradient de domestication, incluant des espèces allant du sauvage au domestique. Cette approche nous permet d’étudier les mouvements de pathogènes entre différentes espèces dans divers environnements. Le Spillover Interface Project, mené par le Dr Serge Morand à l’université de Kasetsart à Bangkok, Thaïlande, est un projet collaborateur de BCOMING, qui étend l'étude en incluant une zone de reforestation, anciennement agricole qui n’est plus exploité et retrouve un état plus naturel, et en explorant plus en profondeur l'interface homme-animal. Le choix de Salmonella spp. comme objet d'étude est pertinent compte tenu de la connaissance extensive de cette bactérie pathogène. La salmonellose est une infection commune chez les animaux et les humains, responsable de gastroentérites chez l’homme et étant la troisième cause de décès par intoxication alimentaire dans le monde (Ferrari et al., 2019; Majowicz et al., 2010; Smith et al., 2014).
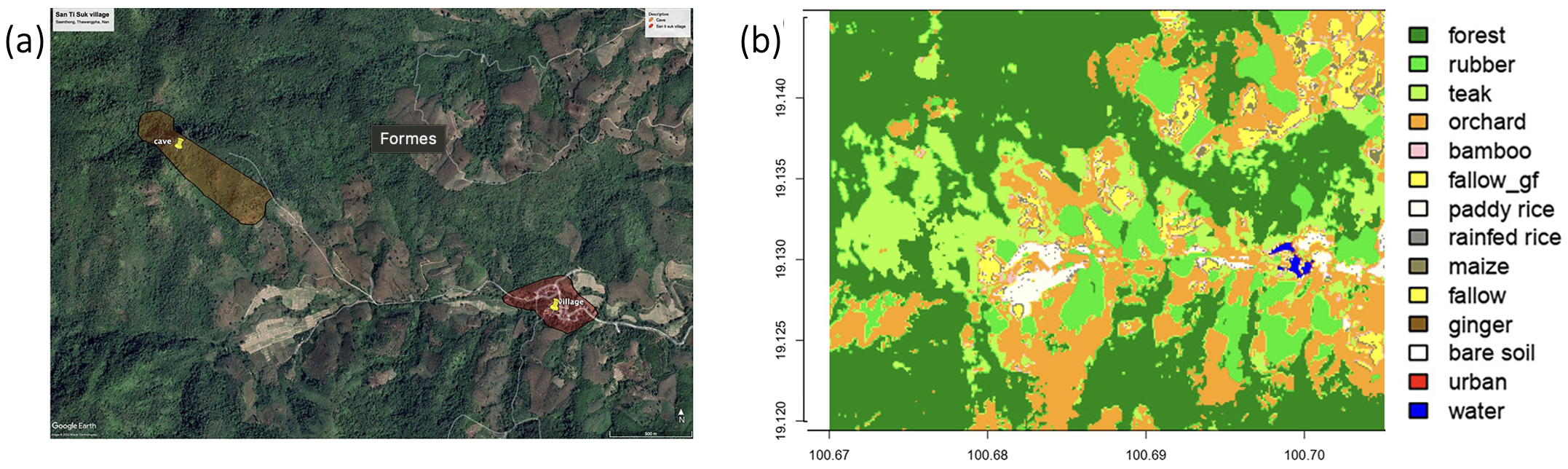
Figure 1 : Extrait de Thinphovong et al., (2024), Localisation de l'étude dans le sous-district de Saen Thong (province de Nan, Thaïlande), précisément (a) dans la partie en altitude du sous-district. Une carte d'utilisation des terres décrit les différentes classes de terres : forêts multispecifiques, plantations (hévéa, teck, plantations de bambou, vergers), cultures en jachère (maïs, riz paddy, gingembre, etc.), infrastructures urbaines, la grotte et le village sont indiqués, (b) la partie en altitude du sous-district.
Notre étude utilise des prélèvements effectués dans la région de Nan en Thaïlande, à proximité de la réserve de Namtanburi. La figure 1 détaille l’environnement dans lequel l’étude a été menée. Nos analyses ont été effectuées sur des échantillons rectaux provenant de chauves-souries, de petits mammifères et de chiens sur plusieurs périodes s'étendant de février 2022 à février 2023. L'ADN a été extrait à l'aide de kits standardisés dans le laboratoire de l’université de Kasetsart à Bangkok, Thailand illustré par les photos 1 et 2. Ensuite, les ADN extraits ont été envoyés en Belgique au GECOLAB et au laboratoire de pathologie du département vétérinaire de l’Université de Liège pour en faire l’analyseUne technique appelée PCR quantitative a été utilisée pour détecter et quantifier spécifiquement Salmonella spp., en se concentrant sur les types dangereux pour l'homme. Cette méthode permet d'amplifier une partie spécifique d'un gène pour détecter l'organisme qui nous intéresse. Enfin, une autre PCR quantitative a été faite pour vérifier que nos échantillons contiennent bien de l'ADN et en quelle quantité. Cela sert de contrôle pour s'assurer que tout fonctionne correctement.
Photos 1 et 2: Analyse laboratoire des échantillons du projet Spillover Interface, Laboratoire de Kasetsart University, Bangkok (Thailand), prise le 29/11/23. Crédit Photo : Pauline Van Leeuwen.
Nos résultats montrent que 12% des échantillons étaient positifs pour Salmonella. Voici la répartition par groupe :
- Chauves-souris (n=45): 20% de positifs, principalement chez Scotophilus heathii, Hipposideros armiger, et Rhinolophus pusillus.
- Petit mammifères (n=48): Aucun des échantillons n'était positif.
- Chiens (n=23): 21,7% de positifs.
Ces résultats sont en partie biaisés par la faible quantité d'ADN dans les échantillons, suggérant une sous-estimation possible des prévalences réelles. En comparaison avec les attentes basées sur la littérature (5% pour les chauves-souris, 7% pour les chiens et 15% pour les rongeurs (Dróżdż et al., 2020; Reyes et al., 2019; Ribas et al., 2016), nos résultats sont élevés pour les chauves-souris et les chiens. Il est surprenant qu’aucun petit mammifère ne soit positif.
L'étude met néanmoins en lumière le risque non-négligeable de transmission de Salmonella dans la région de Nan. Pour réduire ce risque, il est crucial de comprendre les mouvements des animaux et notamment les interactions entre la faune sauvage, les animaux périurbains et les animaux domestiques.
Cette étude souligne l'importance de surveiller les interfaces zoonotiques pour comprendre et essayer de prévenir l'émergence de nouvelles épidémies. La région de Nan, avec sa diversité d'habitats et d'espèces, offre une opportunité unique pour comprendre les dynamiques de transmission des pathogènes. La suite de cette étude préliminaire inclut une analyse des microbiomes sur les mêmes échantillons qui permettra une compréhension plus fine des dynamiques de ces populations par le Dr Pauline Van Leeuwen. Une publication est en cours incluant les résultats de nos deux études conjointes.
Suite à cette recherche, j’ai eu l’honneur de recevoir le prix de l’Institut Vétérinaire Tropical de l’Université de Liège, une reconnaissance qui renforce notre engagement à poursuivre nos efforts pour la santé publique et animale. Je remercie encore les équipes des projets BCOMING et Spillover Interface pour cette opportunité ainsi que ceux qui ont partagé cette belle aventure avec moi.